Finding additional segmentations (WIP)
Many institutions have published their own segmentations as NIfTI, NRRD or RAW files.
In this table we curate a few segmentations including their source(s), and link (in the files field of the table) to their folders in this repository.
| Name | Organism | Source | Format | Ref. space | Voxel dim. | Dir. | Ontology | Files | Ref |
|---|---|---|---|---|---|---|---|---|---|
| ABA Human Half | H. sapiens | source | NIfTI | MNI152 | 500 μm | RAS | ontology | files | Ding et al. J. Comp. Neurol. 2016 |
| ABA Human Full | H. sapiens | source | NIfTI | MNI152 | 500 μm | RAS | ontology | files | Ding et al. J. Comp. Neurol. 2016 |
| Hammersmith Atlas | H. sapiens | source | NIfTI | MNI152 | 1 mm | LAS | custom ontology | files | Hammers et al. Human Brain Mapping 2003, Gousias et al. NeuroImage 2007, Faillenot et al. NeuroImage 2017, Wild et al. PLoS ONE 2017, |
| HCP MMP 1.0 | H. sapiens | source | NRRD | MNI152 | 500 μm | RAS | custom ontology | files | Glasser et al. Nature 2016 |
| ABA Mouse CCF2017 | M. musculus | source | NRRD | CCFv3 | 50 μm | PIR | ontology | files | Lein et al. Nature 2007 |
| Drosophila JRC2010 | D. melanogaster | source | NRRD | JRC2010 | 0.62 μm | RAS | custom ontology | files | Bogovic et al. PlOS ONE 2020, Ito et al. Neuron 2014 |
| Chimpanzee Davi130 Juna.Chimp | P. troglodytes | source | NIfTI | Juna.Chimp | 500 μm | LAS | custom ontology | files | Vickery et al. eLife 2020 |
In this section we present some curated segmentations that have
already been processed through coldcuts and are available
as segmentation class objects.
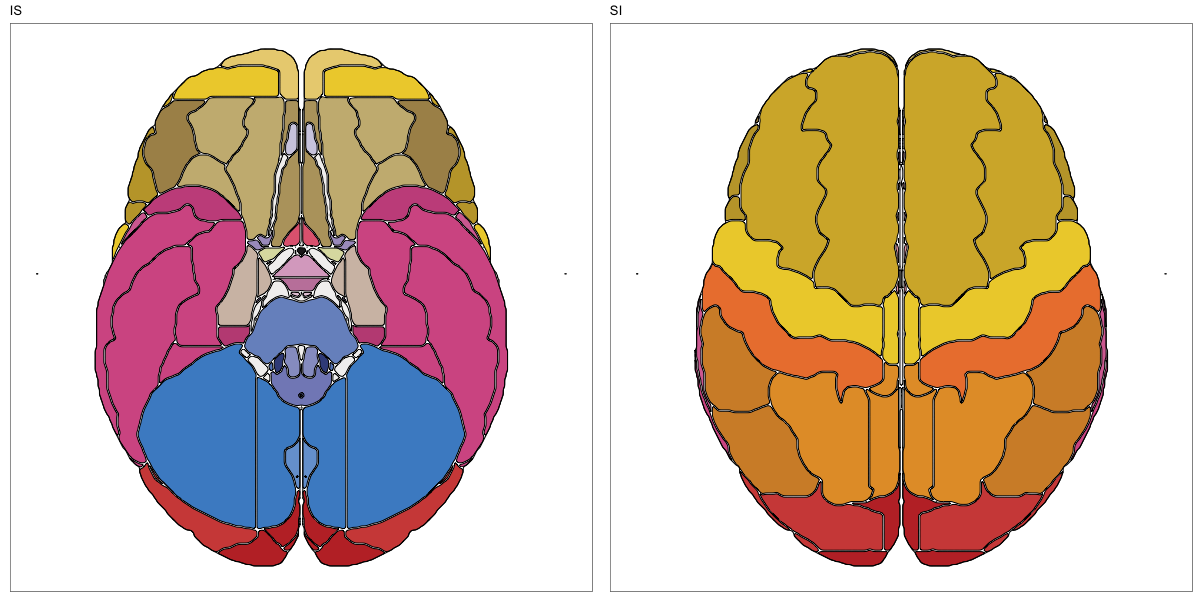
ABA Human (half and full)
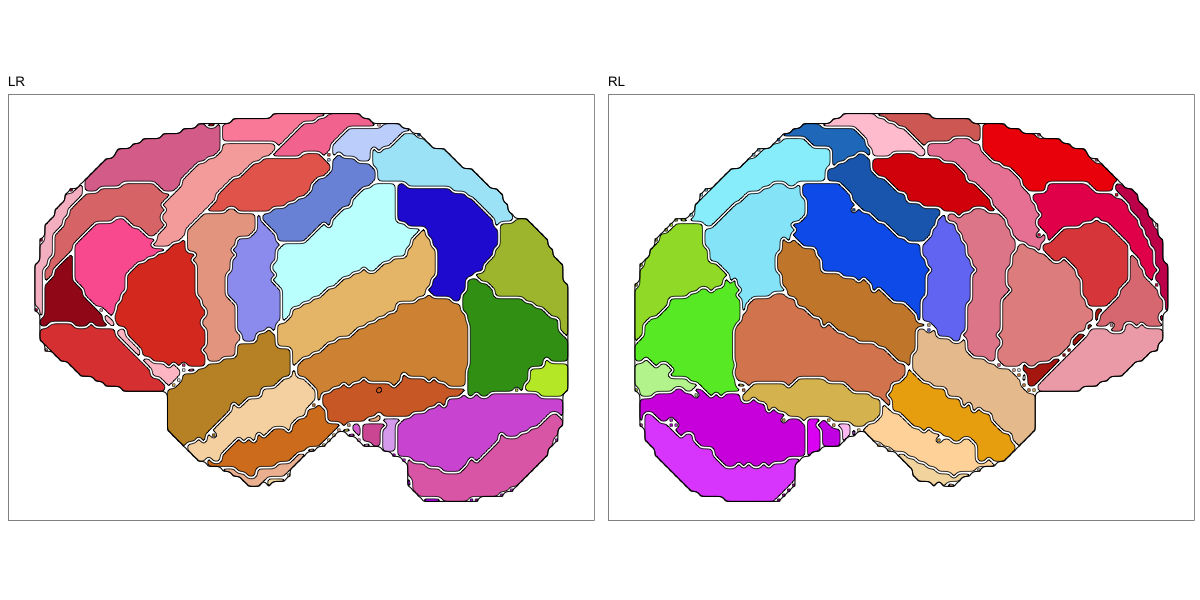
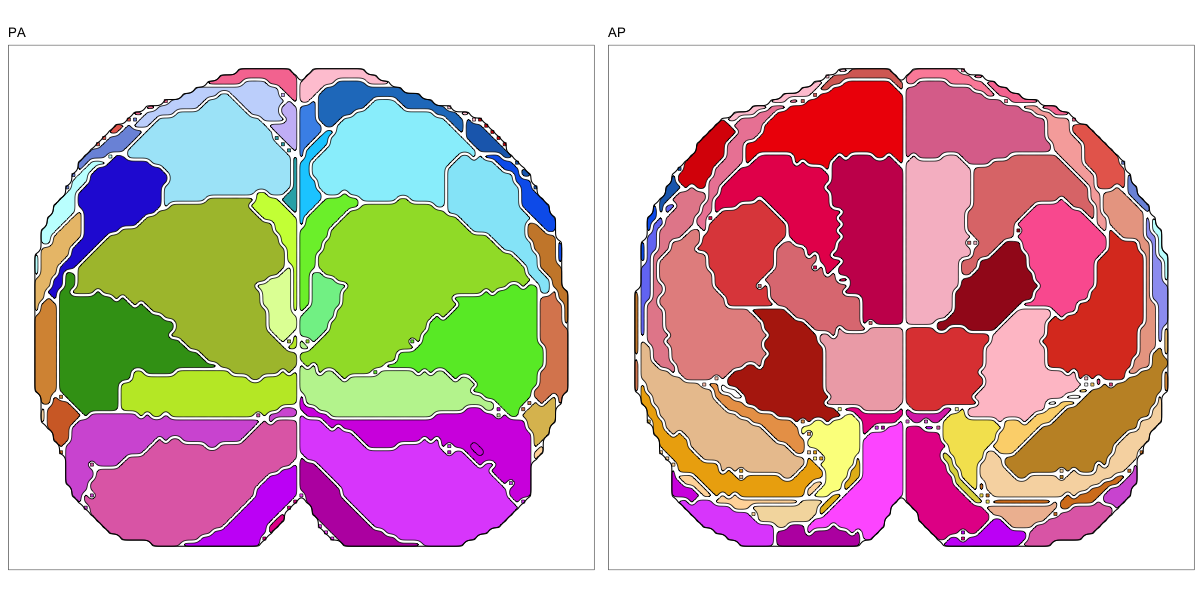
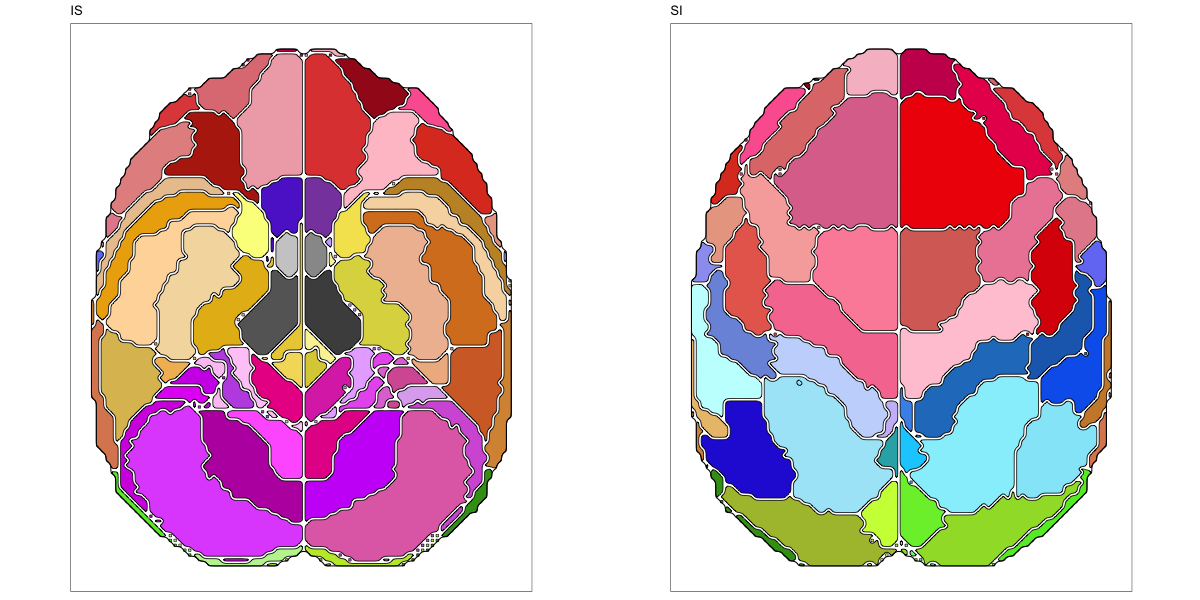
This segmentation was released by the Allen Institute as a volume file for either half emisphere or the full cerebrum, registered to the MNI152 reference space. It is based on the work by Ding et al.
It contains 141 annotated structures, both subcortical and cortical. The structure ontology was provided by the Allen Institute, and the segmentation follows the original ABA color scheme.
The RDS file contains the segmentation class object,
with maximum projections for all structures in all planes, and 3D
meshes.
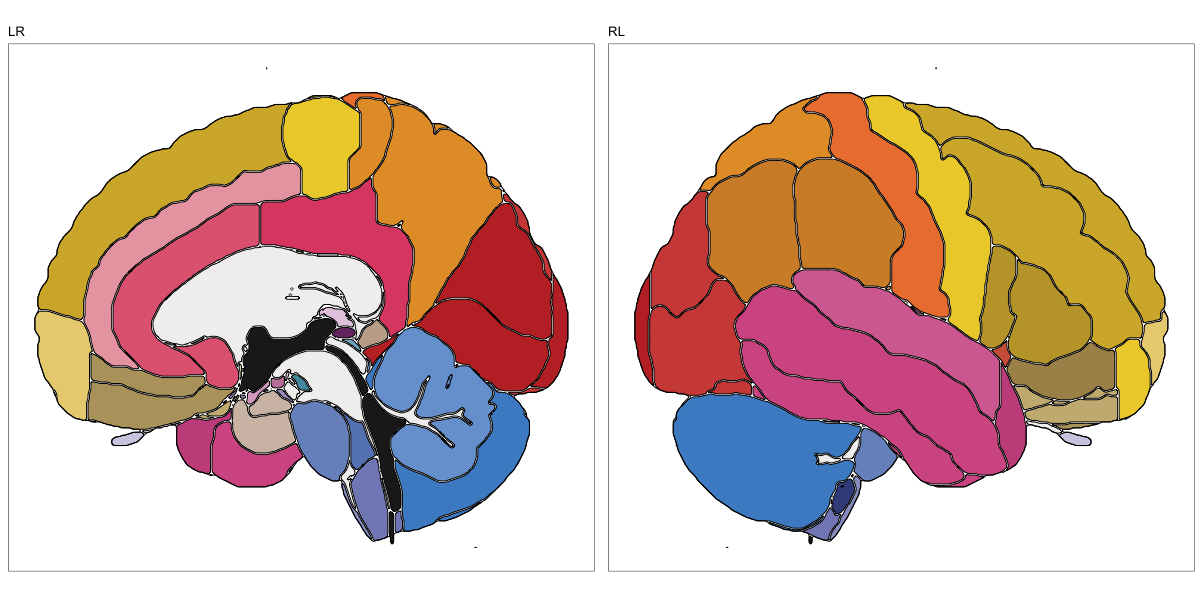
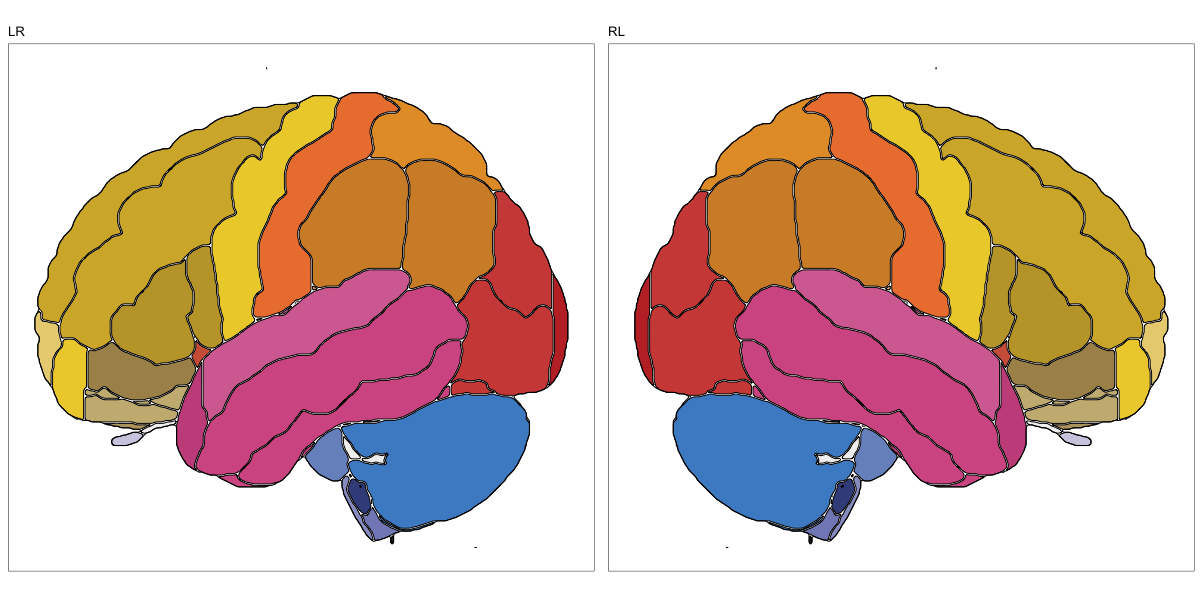
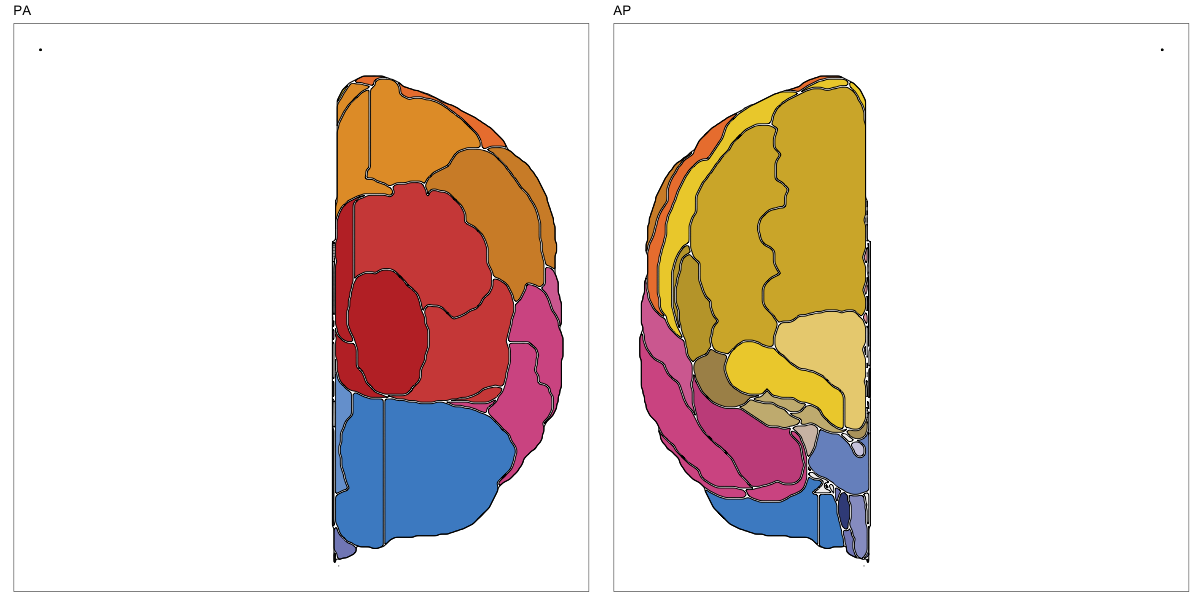
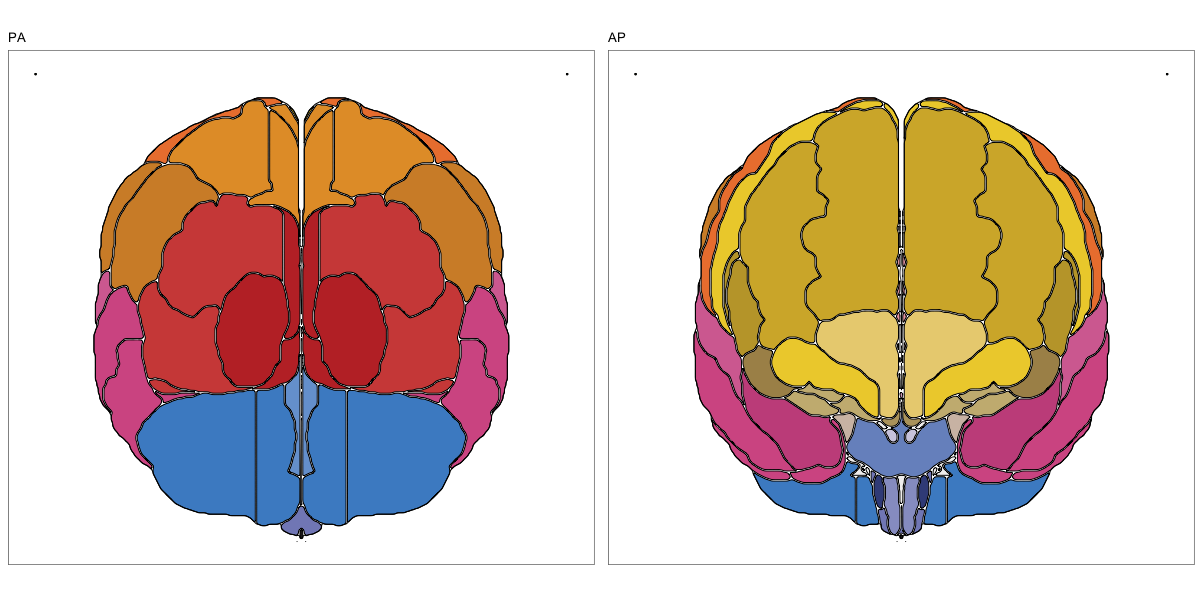

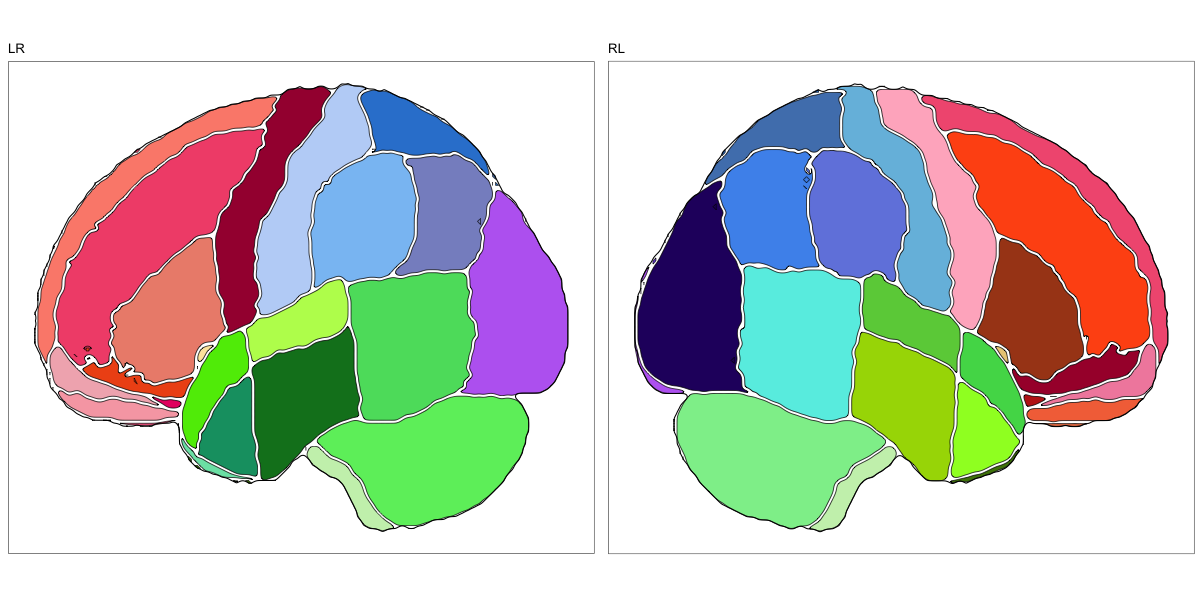
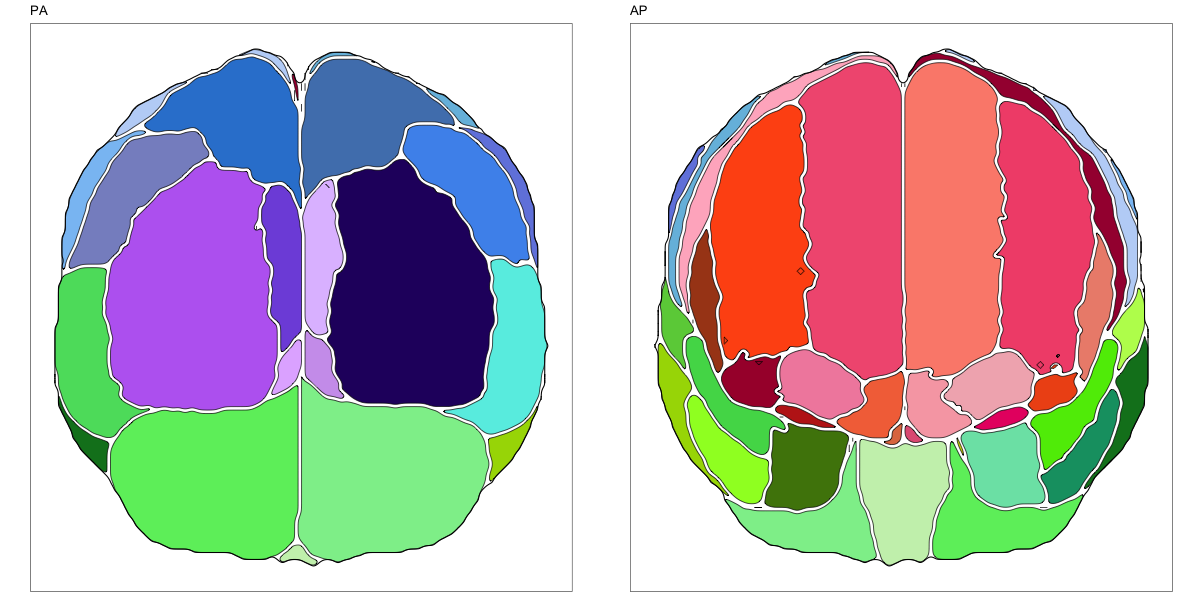
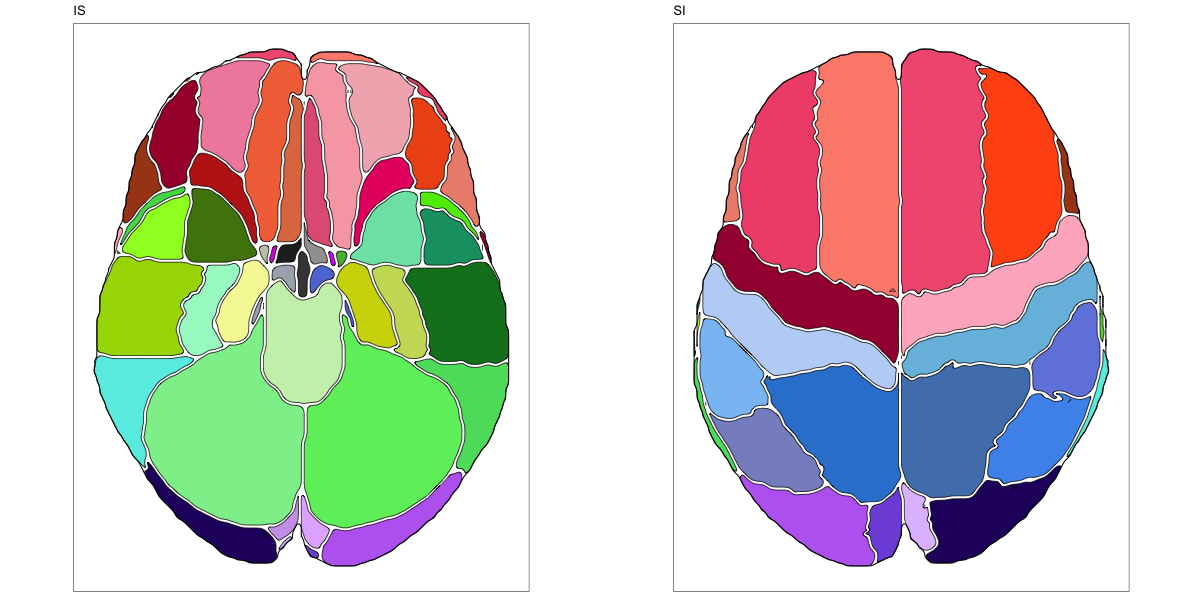
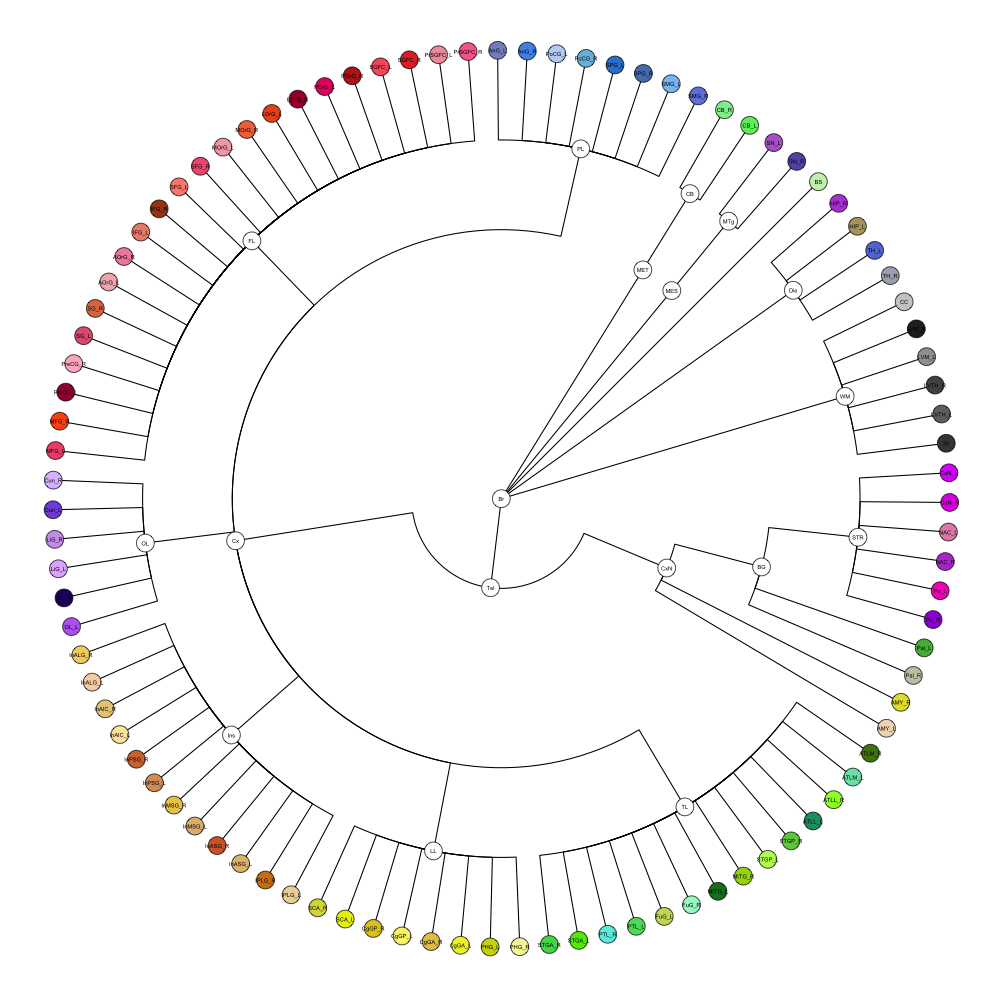
Hammersmith Atlas
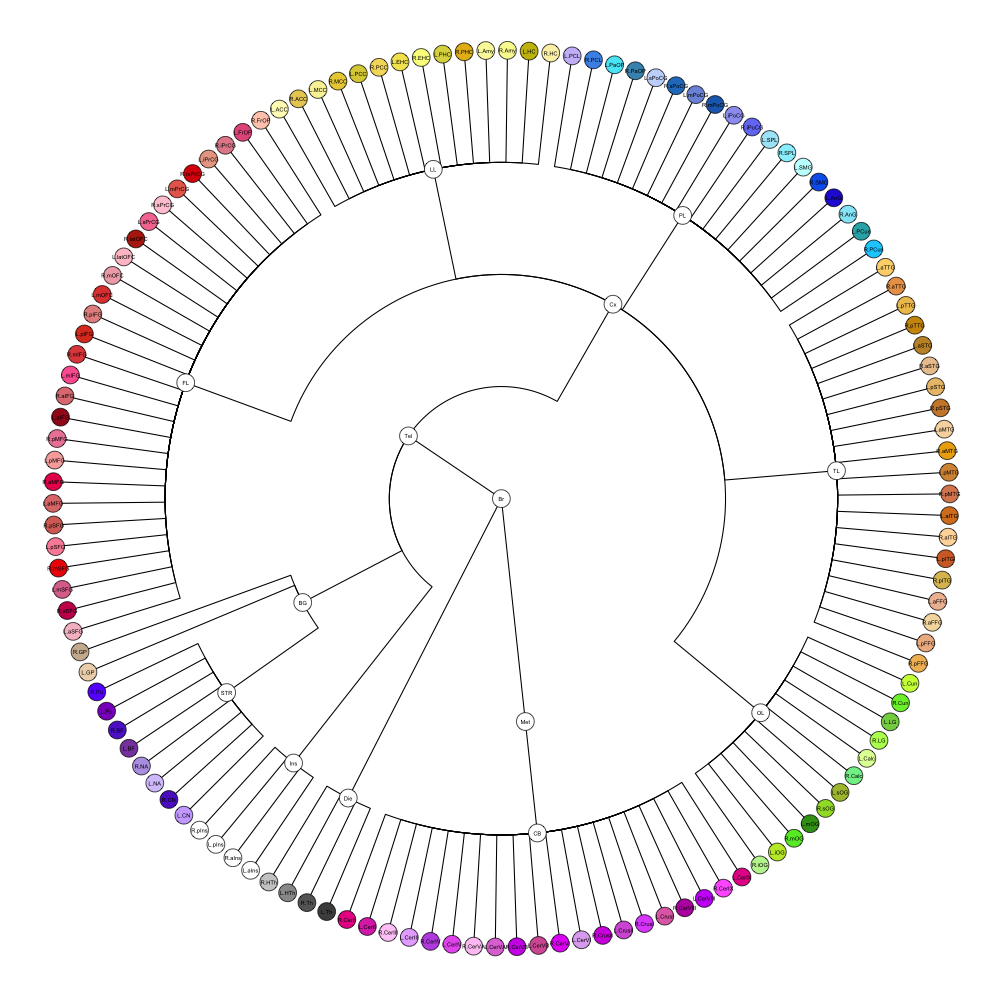
This segmentation was released by the Computational BioImage Group at Imperial College London as a volume file and is based on the work by Hammers et al., Gousias et al., Faillenot et al., and Wild et al. It contains the full cerebrum, registered to the MNI152 reference space. The volume is originally LAS oriented, but was rotated to RAS before drawing. .
It contains 95 annotated structures, both cortical and subcortical, divided in left and right. The structure ontology is custom, largely based on the Supplementary Table A2 from Yaakub et al. 2020 and grouped according to lobes and brain developmental origin. The segmentation follows a “hierarchical random” color scheme.
The RDS file contains the segmentation class object,
with maximum projections for all structures in all planes, and 3D
meshes.
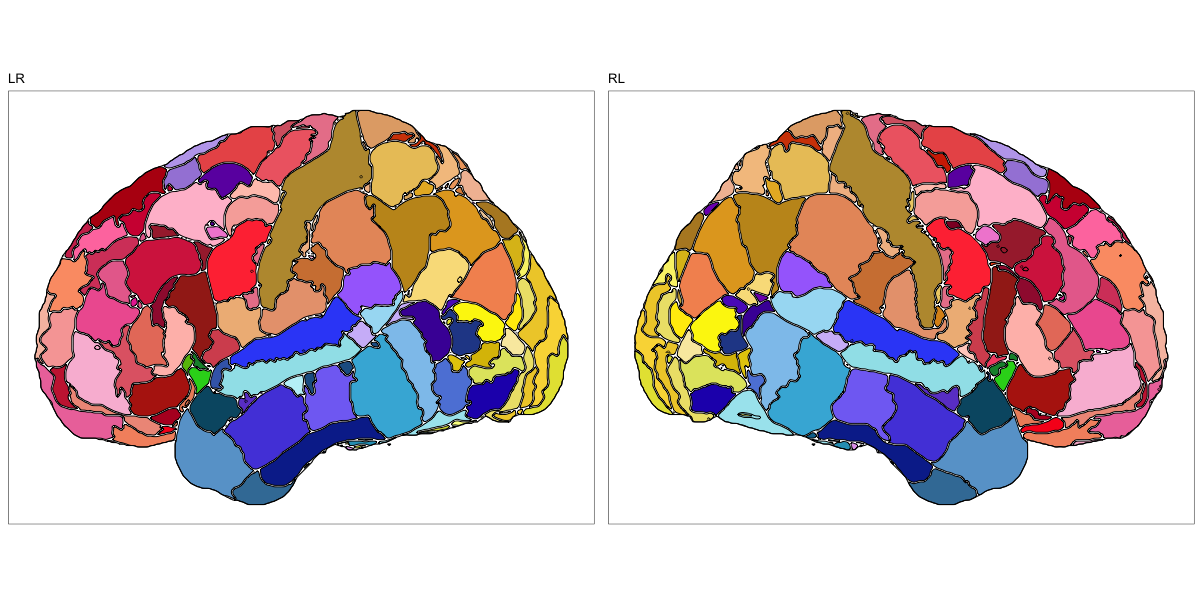
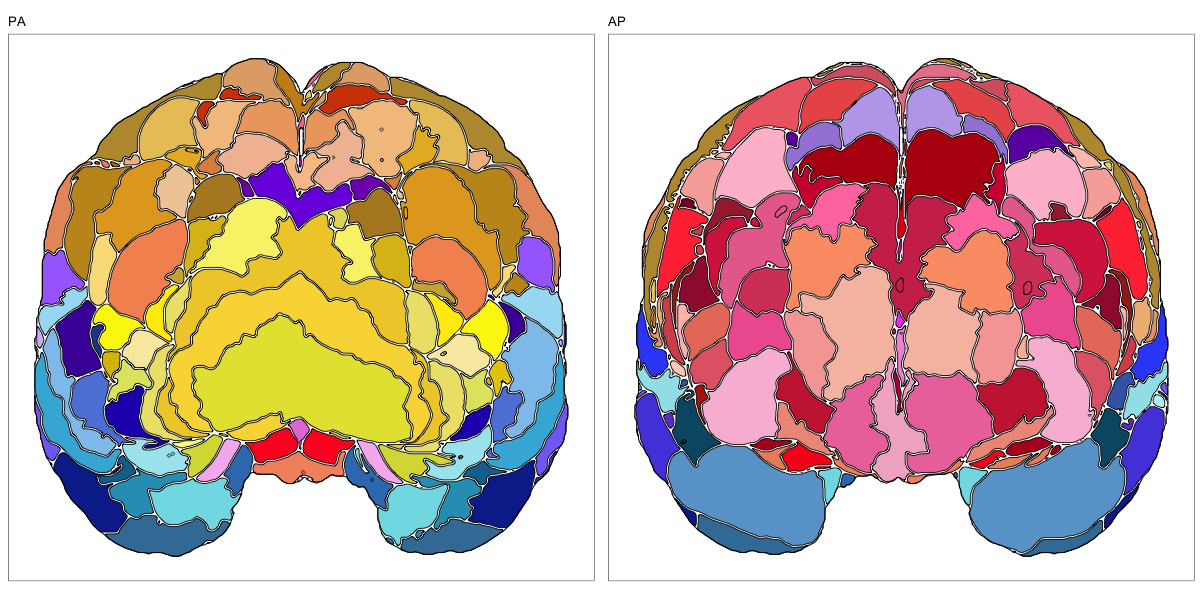
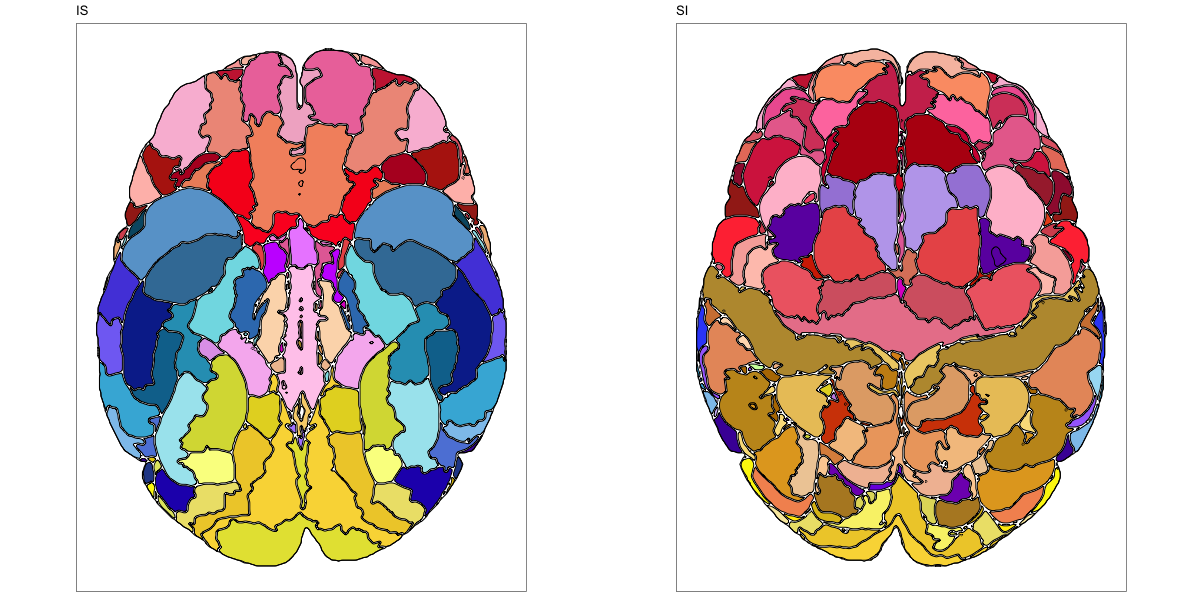
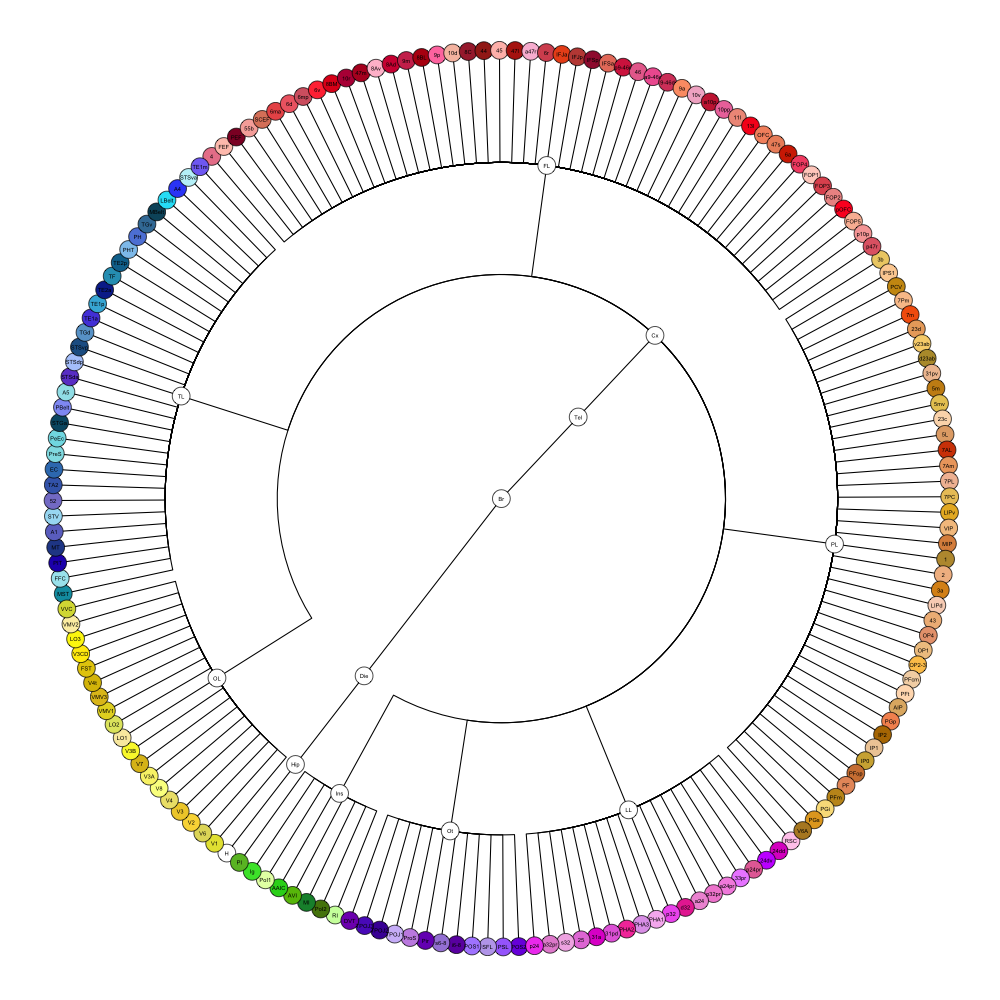
HCP MMP 1.0
This segmentation was released by the Department of Neuroscience, Washington University Medical School, St. Louis, MO, USA as a volume file and is based on the work by Glasser et al. It contains the full cerebrum, registered to the MNI152 reference space. As explained here it should be used with care.
It contains 180 annotated structures, only cortical. The structure ontology is custom, largely based on the Supplementary Table A2 from Glasser et al. and grouped according to lobes and brain developmental origin. Please note that custom ontologies may not be correct in their higher order grouping. The segmentation follows a “hierarchical random” color scheme.
The RDS file contains the segmentation class object,
with maximum projections for all structures in all planes, and 3D
meshes.
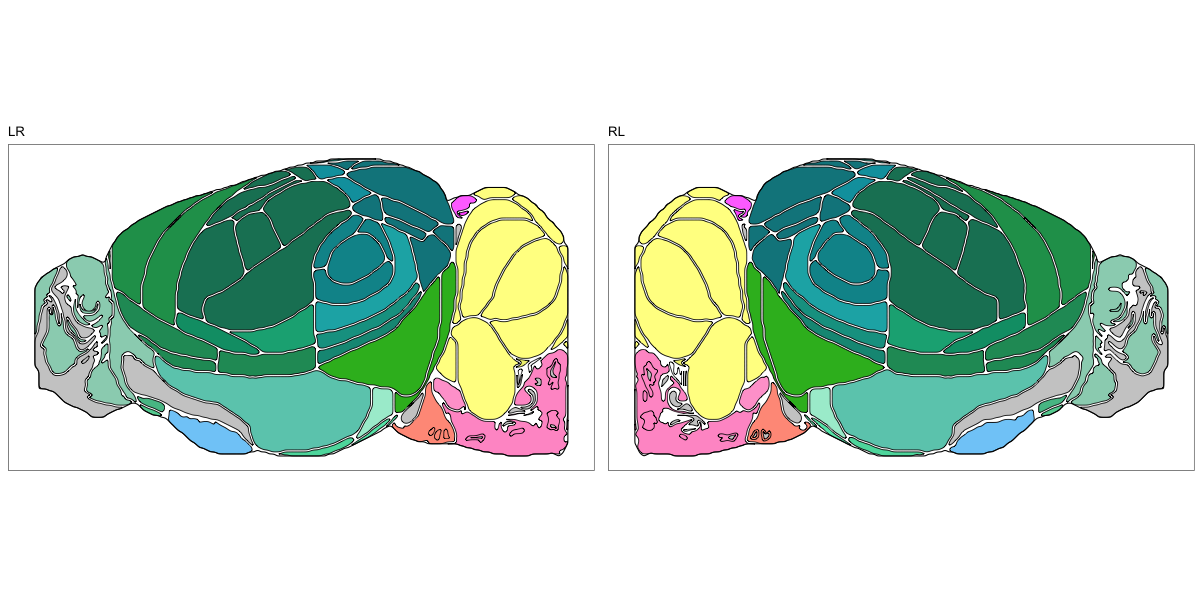
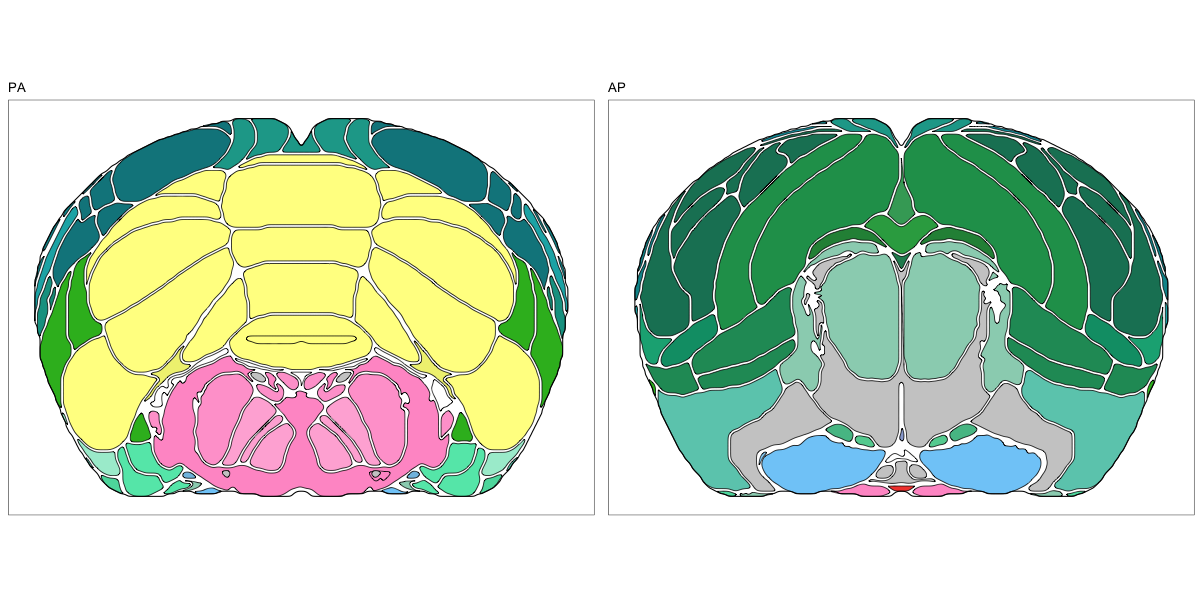
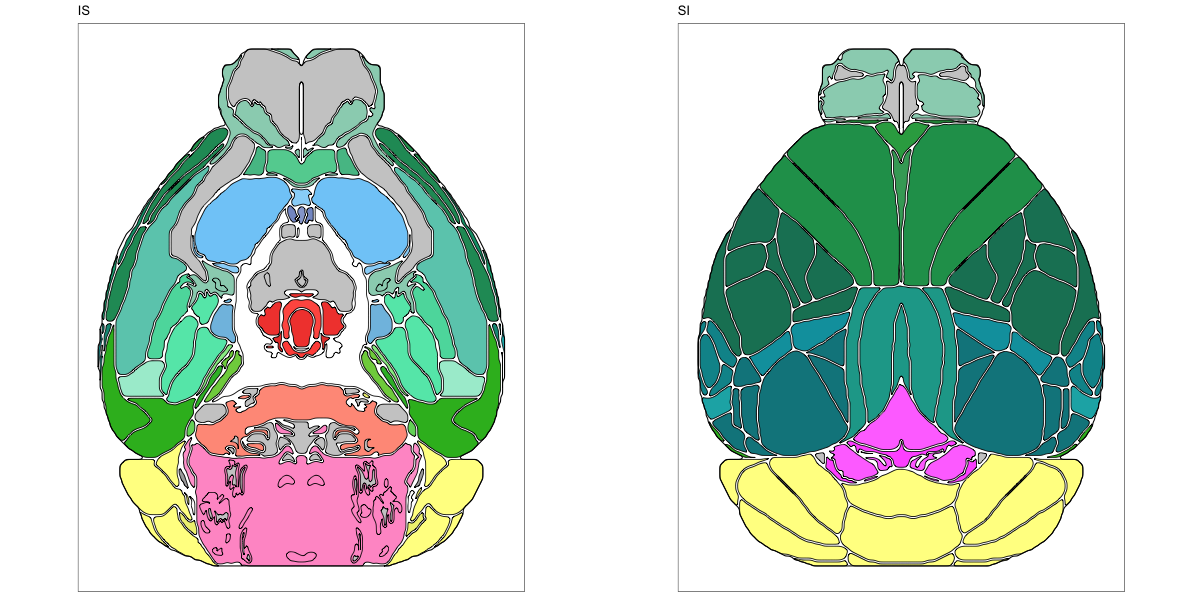
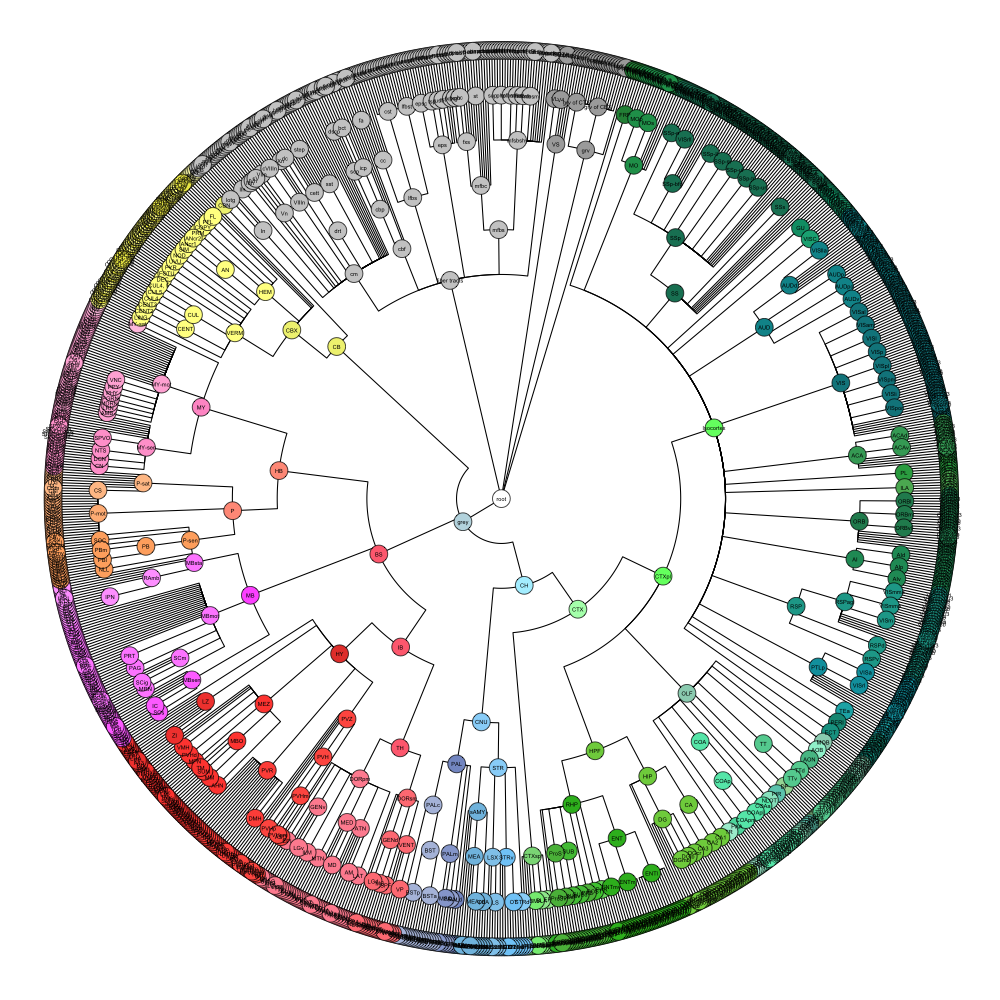
ABA Mouse CCF2017
This segmentation was released by the Allen Institute for Brain Sciences, Seattle, WA, USA as a volume file, registered to the CCFv3 reference space. It is based on the work by Lein et al.
It contains 670 annotated structures, both cortical and subcortical. The structure ontology was provided by the Allen Institute, and the segmentation follows the original ABA color scheme. The original volume is PIR oriented, but was rotated to RAS before drawing.
The RDS file contains the segmentation class object,
with maximum projections for all structures in all planes, and 3D
meshes.
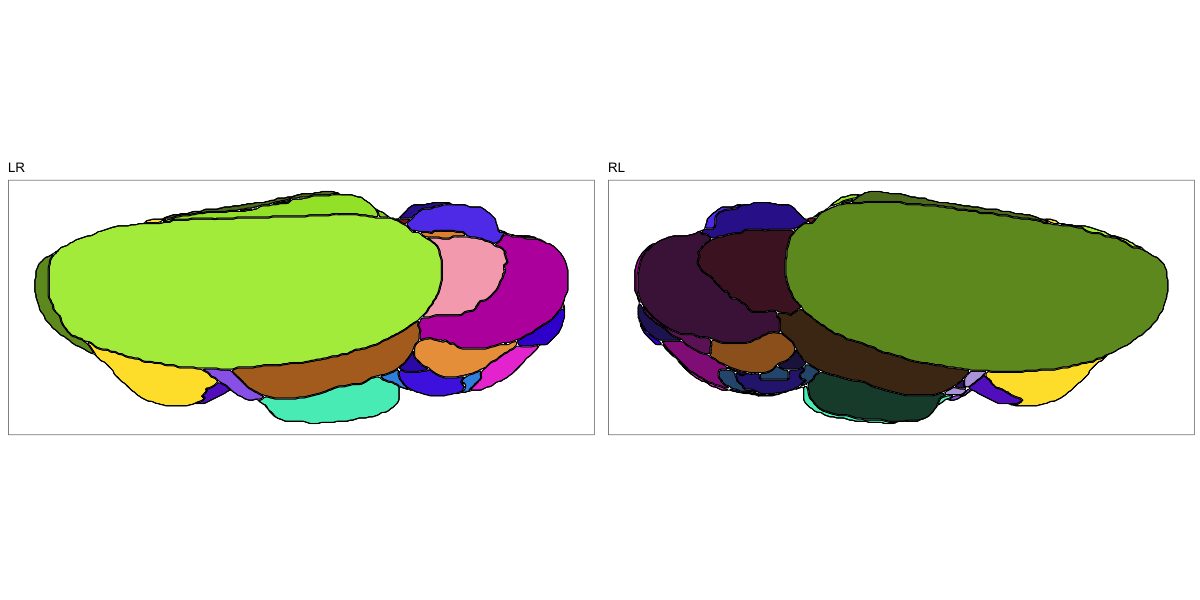
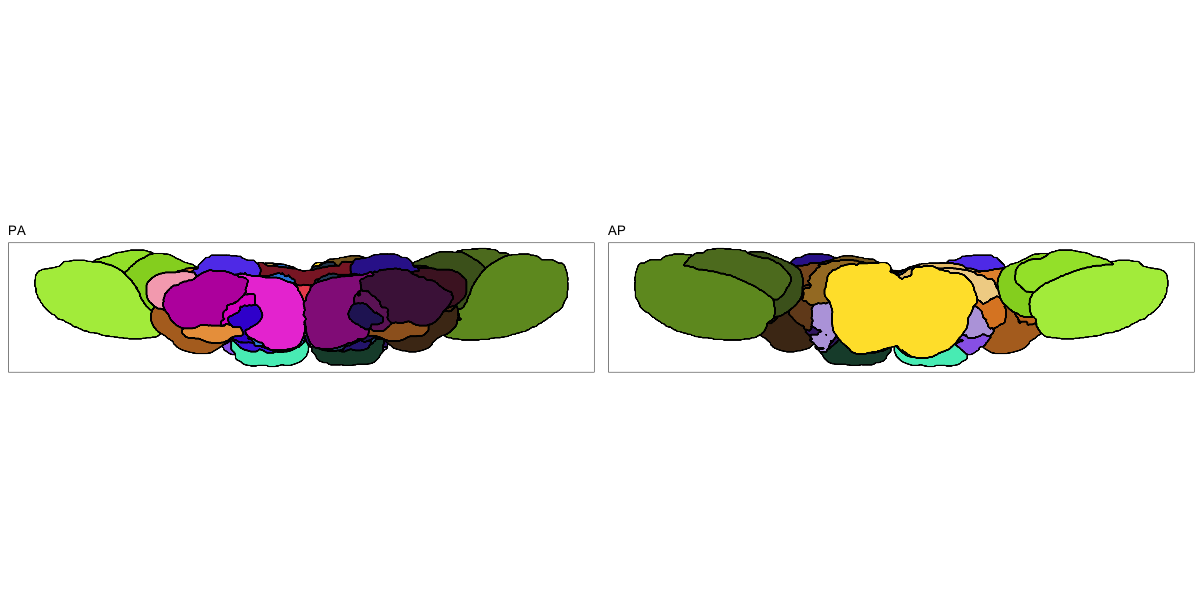
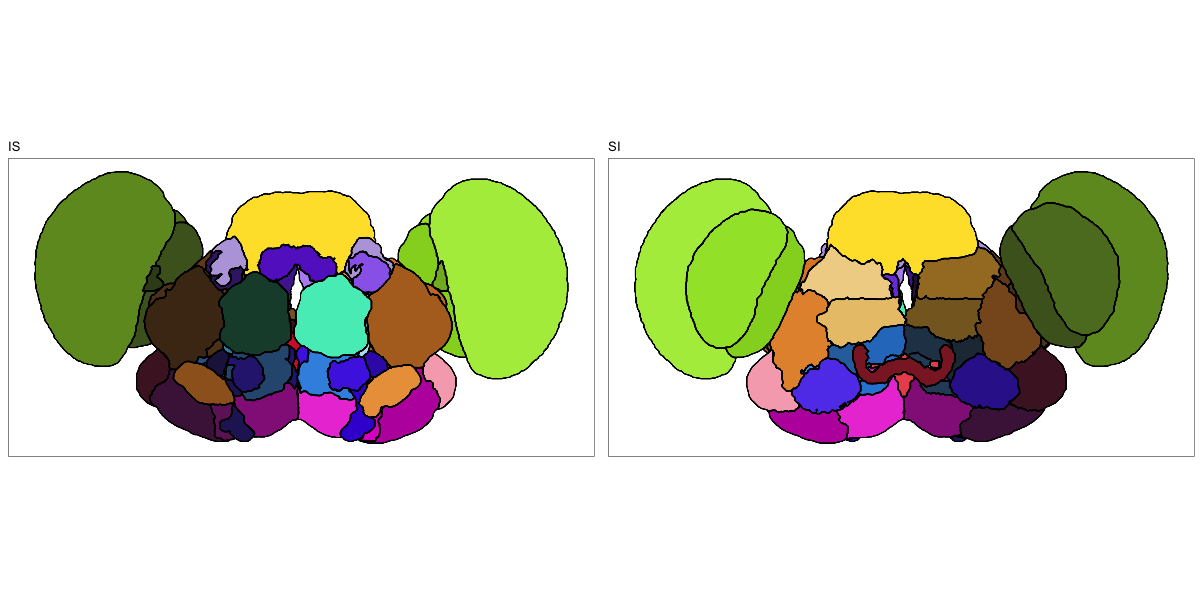
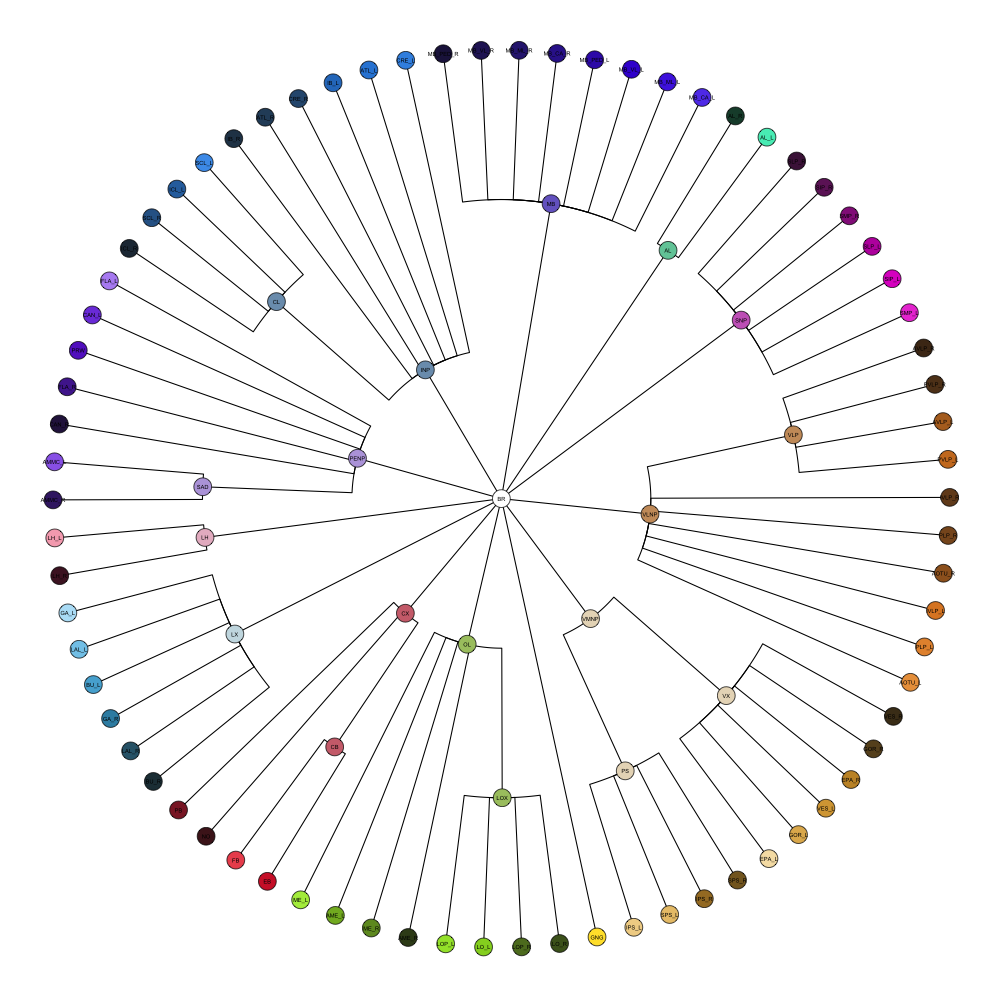
Drosophila JRC2010
This segmentation was released by the Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA, USA as a volume file, registered to the JRC2010 reference space. It is based on the work by Bogovic et al. and Ito et al.
It contains 75 annotated structures, divided in left and right. The structure ontology is custom, largely based on the Table 1 from Ito et al. Please note that custom ontologies may not be correct in their higher order grouping. The segmentation follows a “hierarchical random” color scheme.
The RDS file contains the segmentation class object,
with maximum projections for all structures in all planes, and 3D
meshes.
 /p>
/p>
Chimpanzee Davi130 Juna.Chimp
This segmentation was released by the Institute of Neuroscience and Medicine (INM-7) Research Centre Jülich, Jülich, Germany as a volume file, registered to the Juna.Chimp reference space. It is based on the work by Vickery et al. It contains 130 annotated structures, divided in left and right. The structure ontology is custom, largely based on Figure 2 from Vickery et al. Please note that custom ontologies may not be correct in their higher order grouping. The segmentation follows a “hierarchical random” color scheme. The original volume is LAS oriented, but was rotated to RAS before drawing.
The RDS file contains the segmentation class object,
with maximum projections for all structures in all planes, and 3D
meshes.